Transcriptomics-based longevity drug discovery
Contents
Transcriptomics-based longevity drug discovery#
This project aims to reproduce treatment-screening procedure developed by Georges Janssens et al. [Janssens et al., 2019].
Background#
Aging is associated human morbidity and mortality and there are no way to revert it. However, scientists search for treatments that can slow down aging. One of the approaches was proposed by Georges Janssens et al. in 2019 [Janssens et al., 2019]. Authors used two transcriptomics datasets:
Their own age-stratified RNA-seq data (transcriptome ~ age)
Connectivity Map (CMap) public data (transcriptome ~ treatment)
The key idea is to find treatments that lead to a “younger” transcriptome.
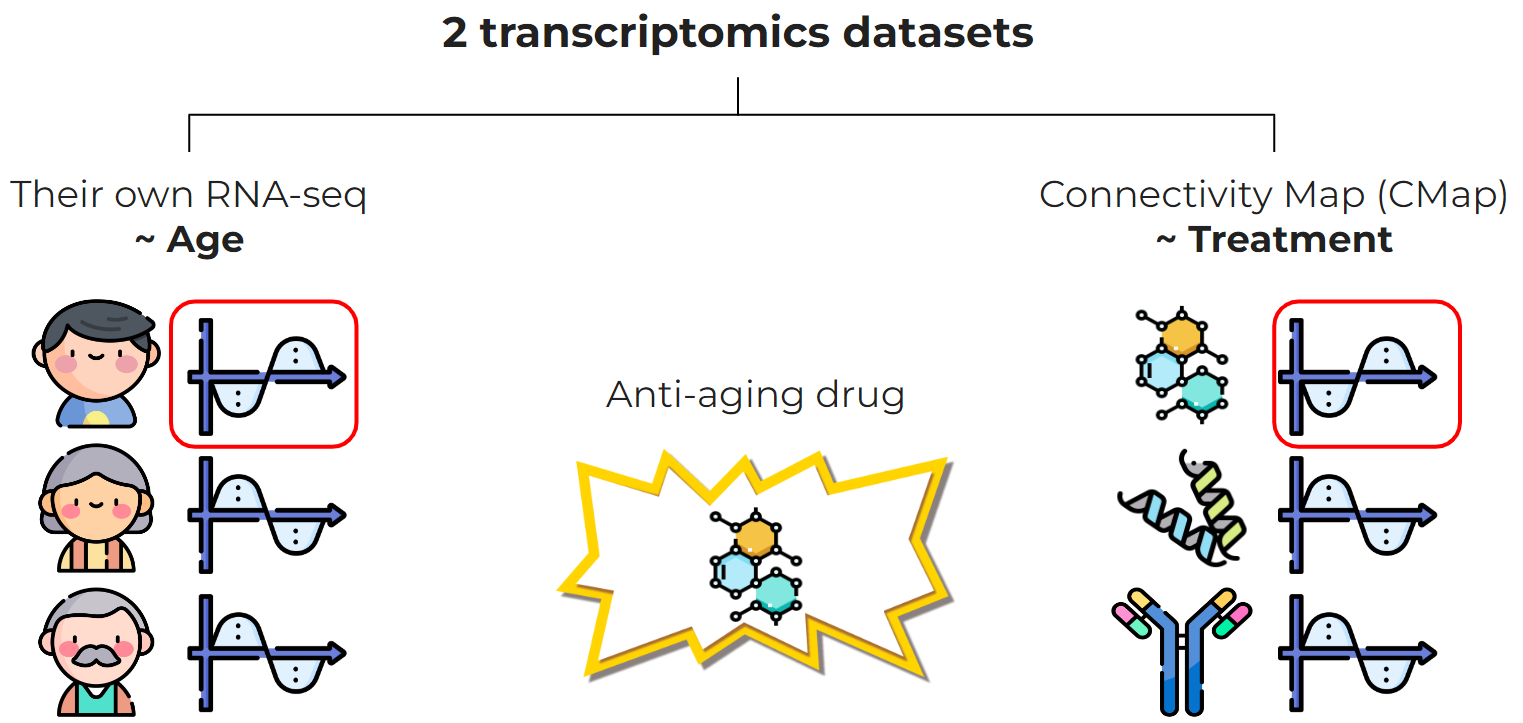
Fig. 15 The overall schema of transcriptome-based treatments selections#
Authors found several candidate treatments and specifically discuss some of them. For instance, they hightlight tanespimycin and monodren involved in Hsp90 inhibition pathways. Hsp90 inhibition was discussed as a process that may be a potential target in developing anti-aging interventions. Janssens et al. also performend in vivo experiments in C. elegans that support their findings.
Materials and methods#
All data processing was performed using R v.4.2.3 with tidyverse v.2.0.0 and python v.3.11.6 with pandas v.2.1.2.
GTEx dataset preprocessing
Age-stratified RNA-seq data (GTEx dataset) was downloaded from the GTEx portal (”Bulk tissue expression” section). We used already preprocessed gene RPKM data from the GTEx Analysis V6p release. We also obtained metadata description from the “Metadata” section (GTEx Analysis V6p Sample Attributes and Subject Phenotypes). Both annotation files were joined on sample IDs to get combined metadata. Samples were annotated as “young” for ages 20-59 years old and “old” for 60-69 years old.
Based on the metadata, GTEx dataset was divided into several parts based on tissue, age and gender parameters. For the further analysis only brain, heart, kidney, liver and stomach data was selected in order to reduce computational time. Also, for all of the subsets, we retained only transcripts present in both GTEx and CMap datasets (based on the transcript IDs). We also revomed low abundant transcripts with RPKM values less than 0.1 in at least 80% of samples. Additional filtering was applied to keep transcripts with RNA integrity number greater or equal to 6. Resulting transcripts were added a pseudocount and log2-transformed. Then we performed differential expression analysis between “young” and “old” samples. Only differentialy expressed transcripts with p-value less then 0.01 were selected for the further analysis.
CMap dataset preprocessing
Connectivity (CMap) dataset was kindly provided by Dmitrii Kriukov. Control group of CMap dataset was aggregated by cell type to compute cell-wise means. After that we computed log2(Fold Change) values for each treatment with respect to the corresponding cell-wise control.
Transcriptom-based treatment selection
Transcriptom-based treatment selection using machine learning was performed in R v.4.2.3 with caret v.6.0-94, randomForest v.4.7-1.1, ROCR v.1.0-11, tidyverse v.2.0.0, matrixStats v.1.2.0.
Firstly, we built RandomForest model to predict age category (“young” / “old”) based on the preprocessed GTEx transcriptomics data. We built different models for different tissues and selected those with AUC score greater then 0.75 (based on 10-fold cross-validation).
Secondly, we used selected models to find geroproctive treatments. We obtained “middle-aged” transcriptome by averaging “young” and “old” GTEx data. Then we multiplied this “middle-aged” values by the corresping fold changes from the CMap dataset, wich resulted us “drug-induced” transcriptome. Then we made age predictions based on our drug-indiced transcriptome and selected those for which “young”-age probability was higher then for the “old”-ade. We also calculated geroprotective index (GI) by normalization and centering probabilities of “young” age class (PY):
The results were visualised in python v.3.11.6 with pandas v.2.1.2, matplotlib v.3.8.0 and seaborn v.0.13.0.
Results#
We built 5 transcriptome-based age prediction models for each of the selected organs (brain, heart, kidney, liver and stomach). They were applied to the 100 randomly selected treatments and only brain model resulted in one geroprotective compound - L-ergothioneine. Then we used brain model as a more promising one and applied it to all 3546 treatment in the CMap dataset.
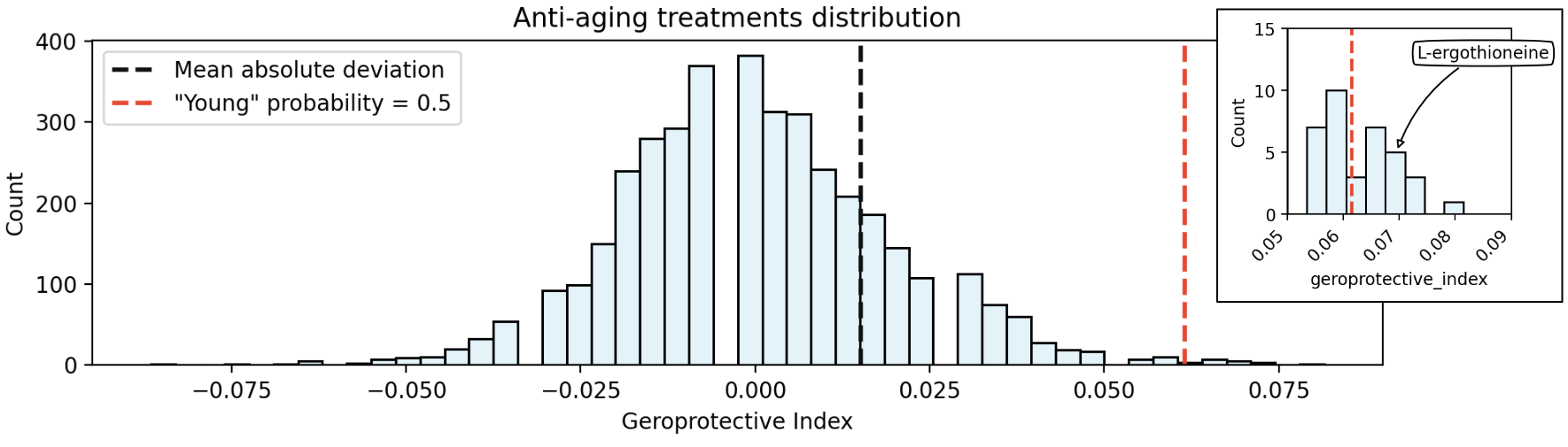
Fig. 16 Geroprotective index distributions of all treatments. Black line depicts selection threshold in the original paper (however authors also used some ranking procedure)), red line represents our threshold (probability of “young” age is equal to 0.5).#
Only 11 compounds were found to show geroprotective results: olanzapine, XL-888, ritonavir, thioguanine, L-ergothioneine, nafamostat , ochratoxin-a, etofylline-clofibrate, tebipenem , E-2012, EMF-sumo1-7. Among them, thioguanine showed the most geroprotective index (0.0815), and the olanzapine, XL-888, ritonavir had the secong highest GI (0.0740). L-ergothioneine had the third highest GI (0.0670), but we see this molecule as the most promising as it was present in the results of several analyses.
| Probability | Treatment | Geroprotective Index | |
|---|---|---|---|
| "Old" | "Young" | ||
| 0.498 | 0.502 | EMF-sumo1-7 | 0.0656 |
| 0.496 | 0.504 | ochratoxin-a | 0.0670 |
| 0.498 | 0.502 | nafamostat | 0.0656 |
| 0.496 | 0.504 | L-ergothioneine | 0.0670 |
| 0.494 | 0.506 | olanzapine | 0.0740 |
| 0.494 | 0.506 | XL-888 | 0.0740 |
| 0.498 | 0.502 | E-2012 | 0.0656 |
| 0.494 | 0.506 | ritonavir | 0.0740 |
| 0.49 | 0.51 | thioguanine | 0.0815 |
| 0.498 | 0.502 | tebipenem | 0.0656 |
| 0.496 | 0.504 | etofylline-clofibrate | 0.0670 |
Discussion#
In this project, we built a Random Forest model for the age classification of transcriptomic data and applied it to find potential anti-ageing treatments. Our model identified 11 compounds that could potentially be geroprotective. However, high machine learning metrics are not a sufficient condition for the quality of the results. To validate the results, we additionally analysed candidate molecules. We selected the top 4 molecules according to the geroprotective index (thioguanine, olanzapine, XL-888, ritonavir), also L-ergothioneine, as it was supported by several runs of the analysis.
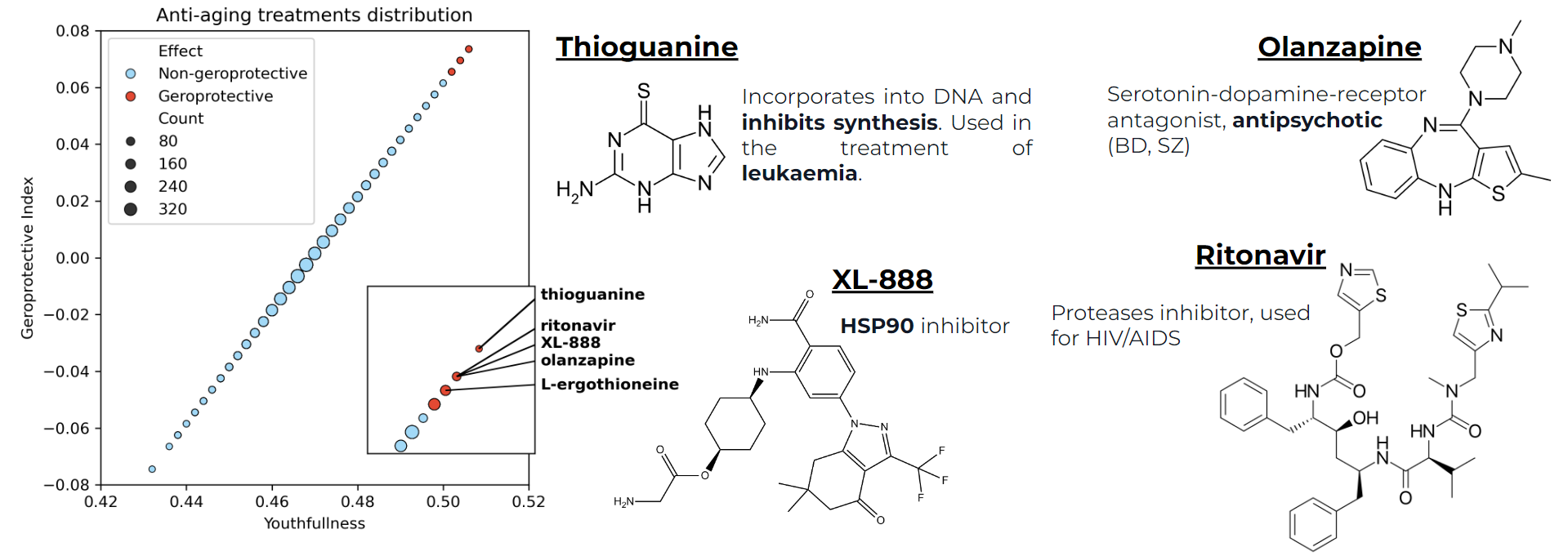
Fig. 17 Geroprotective index distributions of all treatments and description of top-4 treatments with respect to the geroprotective index (thioguanine, olanzapine, XL-888, ritonavir).#
Geroprotective molecules overview#
Among the isolated molecules, most of them are already used drugs in one or another field.
Thioguanine, the top 1 molecule in the geroprotective index, is 2-aminopurine. Its key action is incorporation into DNA, resulting in inhibition of synthesis and replication. Therefore, it is actively used in the therapy of acute leukaemia. Most likely it is possible to find evidence for the geroprotective properties of thioguanine, nevertheless, its exact involvement in the mechanism of aging replacement seems a bit vague to us.
Olanzapine is a benzene molecule containing sulphur atoms and a nitrogen antagonist. It acts as a serotonin-dopamine receptor antagonist and is used as an antipsychotic medication (second-generation antipsychotic). It is used for disorders such as schizophrenia and bipolar disorder and is also known as Zyprexa. In this form, the relationship of olazapine to aging may not be completely clear, but in recent years there has been a growing body of work on the geroprotective properties of antipsychotics and antidepressants.
XL-888 is a less studied molecule. The available data suggest that it is an ATP-competitive inhibitor of the chaperone Hsp90. This is an important remark regarding our work, because in the original article the authors elaborate on the mechanism of Hsp90 inhibition as an important target in anti-aging. They identified other inhibitors, but in any case this overlap is very valuable to us.
Ritonavir is an L-valine derivative and also known under the name Norvir. It is an inhibitor of various proteases and in medicine is used as an HIV protease inhibitor to treat AIDS/HIV as well as various forms of hepatitis. In general, it can be a broad-spectrum antiviral drug, but it is often used to boost the effects of other drugs due to its ability to inhibit liver cytochrome P450-3A4.
L-ergothioneine anti-aging treatment#
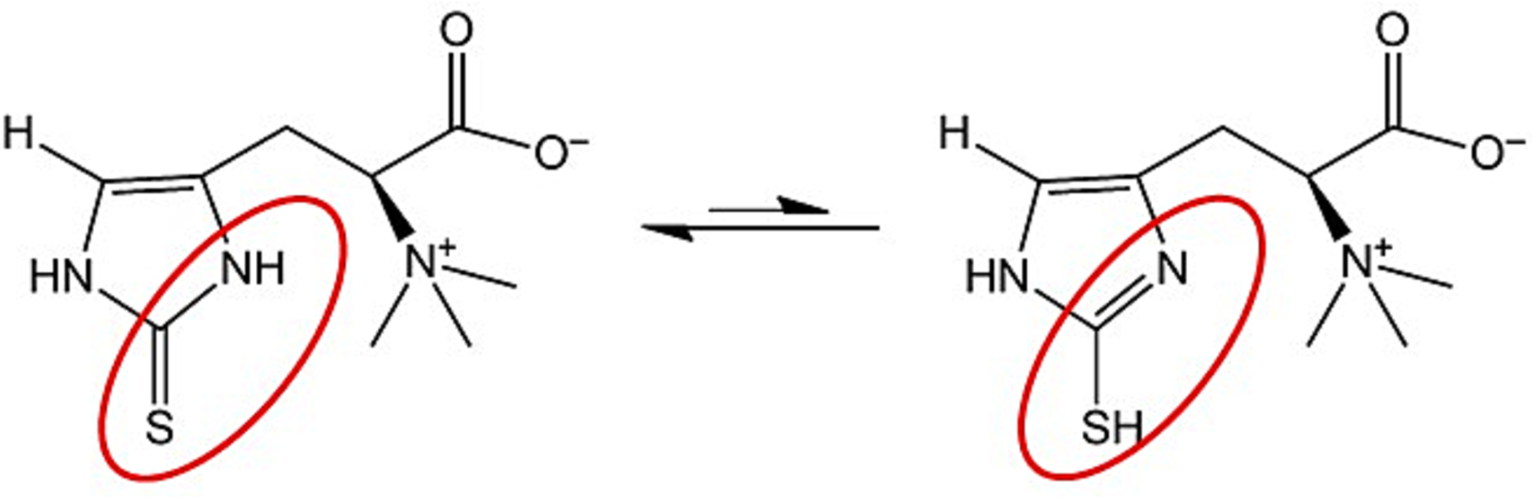
Fig. 18 The chemical structure of the L-ergothioneine, red oval higlights the functional group [Apparoo et al., 2022].#
L-ergothioneine is a histigine-derivative aminoacid with sulphur group on the imidazole ring. This sulphur can be found in the oxidised and reduced forms, which defines it’s function. L-ergothioneine was proposed to have protection function against reactive oxygen species [Apparoo et al., 2022, Cheah et al., 2016, Chen et al., 2023, Paul and Snyder, 2010]. Paul et al. using HeLa cells lines showed that L-ergothioneine increases cells viability against \(H_2O_2\) [Paul and Snyder, 2010]. Therefore, it may be involved in responses to oxidative stress originating from mitochondrial activity, ultraviolet radiation, etc.
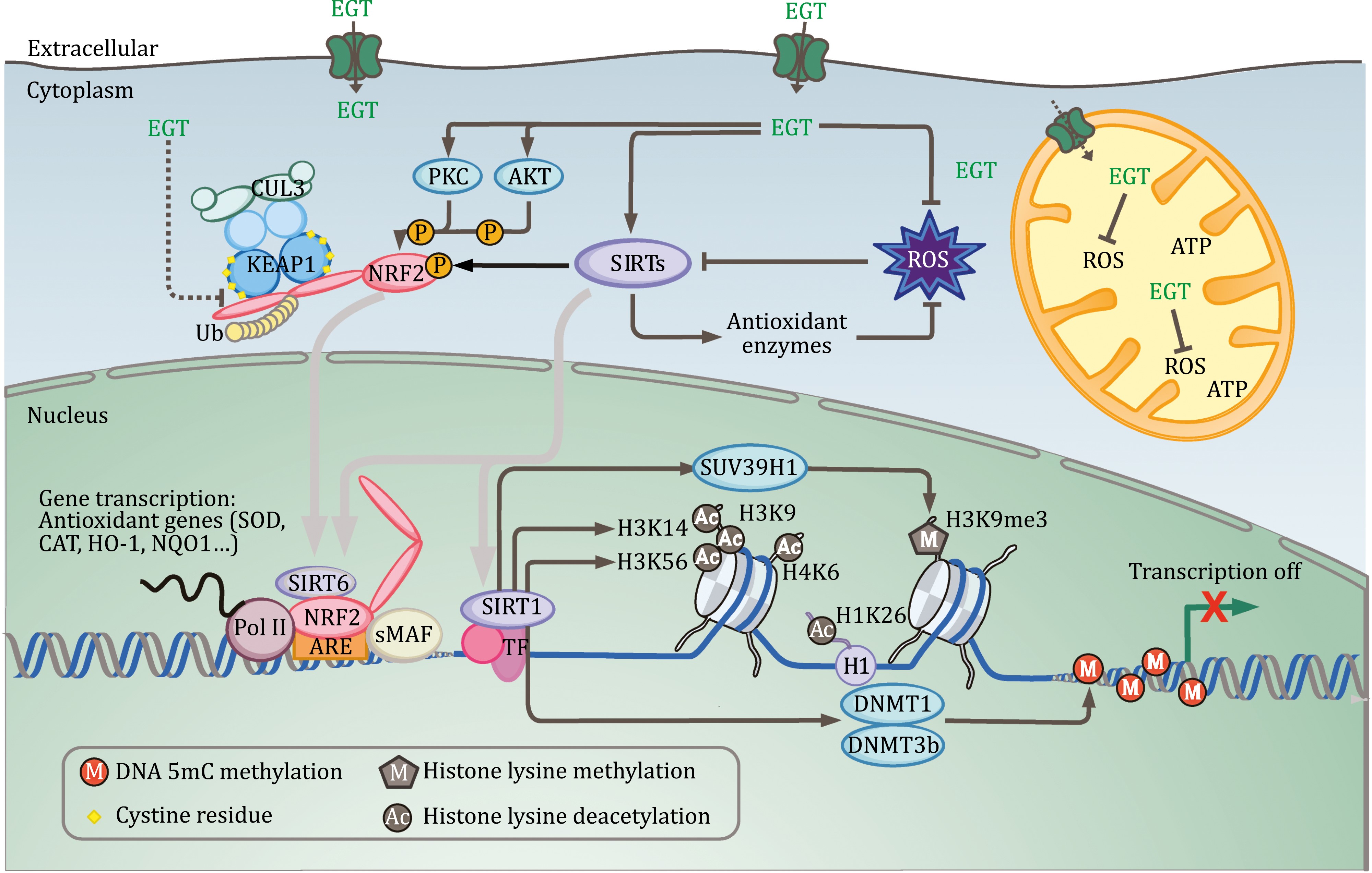
Fig. 19 L-ergothioneine in the network of the hallmarks of aging [Chen et al., 2023].#
These features of L-ergothioneine make it an interesting member of the aging hallmarks network [Apparoo et al., 2022, Chen et al., 2023]. First of all, it is an obvious element of the cell’s antioxidant system. Due to this, it has an immediate influence on the maintenance of genomic stability and mitochondrial functionality. However, there are some other interesting interconnections. For example, L-ergothioneine has been shown to be associated with sirtuin proteins that provide orchestration of the epigenetic landscape in the cell [Chen et al., 2023]. Also an important observation is that L-ergothioneine is not synthesised in the human organism and therefore its maintenance is directly related to the operation of transport systems (in particular OCTN1 channels). Its involvement in various signalling cascades such as MAPK and KEAP1-NRF2 has also been shown.
All this substantiates the role of L-ergothioneine as a geroprotector. Indeed, there is some medical research in this direction. Beelman et al. showed that the amount of L-ergothioneine in the diet is correlated with average mortality. Cheah et al. also reported that the amount of L-ergothioneine in the blood decreases with age. These data give us confidence in the adequacy of the model we have constructed.
Clarification of discrepancies with the findings in the article#
An important issue we would like to address in this discussion is to analyse possible differences with the results in the original paper (see figure below). The first important remark is that among the 11 compounds we found, there are no overlaps with the authors’ results. However, for example, we were able to find other Hsp90 inhibitors, and the mechanism of Hsp90 inhibition is one of the key topics of discussion of potential geroprotectors in the article. Although, the following reasons for the differences can be pointed out:
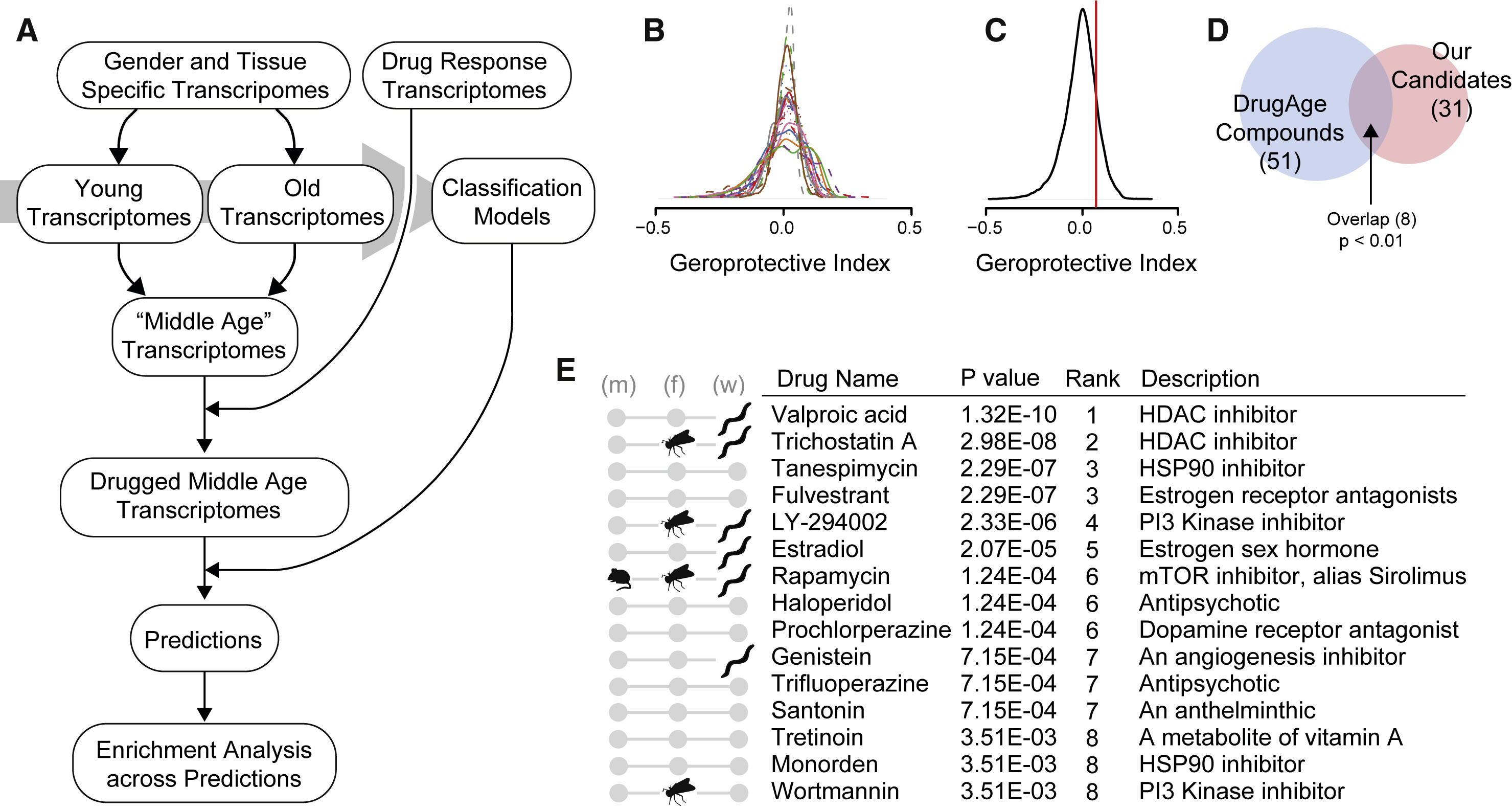
Fig. 20 Anti-aging treatments identified by Janssens et al. (E). The picture is taken from the original paper [Janssens et al., 2019].#
We used reduced data to save time. We selected some organs and ran a sample of 100 treatments (out of 3546 total) on them. We also ran our analysis for all treatments, but only for the brain.
Our compounds-selectiong procedure differs from the paper due to lack of ran models (we did perform ranking)
We suspect we used a slightly different version of CMap dataset. In our data we had 3546 treatments against 1309 in the paper.
Conclusions#
In this project we built transcriptomic age-classification model which was used to identify treatments that result in a “younger” transcriptomes. With this model we identified 11 potentially geroprotective compoings, The most promising of them are: L-ergothioneine, thioguanine, olanzapine, ritonavir, XL-888. These molecules do not match those found by the authors of the original paper, but we discuss possible reasons for this. We also studied L-ergothioneine individually and associate our hopes for a future victory over aging with this very compound.
Credits#
The project was done by:
Ksenia Kubenko preprocessed GTEx and CMap datasets
Ekaterina Kashuk implemened Random Forest models and performed geroprotectors selection
Nikita Vaulin visualised the results, reviewed the found geroprotectors and assisted in the preprocessing
References#
- 1(1,2,3)
Yasaaswini Apparoo, Chia Wei Phan, Umah Rani Kuppusamy, and Vikneswary Sabaratnam. Ergothioneine and its prospects as an anti-ageing compound. Experimental Gerontology, 170:111982, 2022.
- 2
Irwin K Cheah, Lei Feng, Richard MY Tang, Keith HC Lim, and Barry Halliwell. Ergothioneine levels in an elderly population decrease with age and incidence of cognitive decline; a risk factor for neurodegeneration? Biochemical and Biophysical Research Communications, 478(1):162–167, 2016.
- 3(1,2,3,4)
Li Chen, Liping Zhang, Xujun Ye, Zixin Deng, and Changming Zhao. Ergothioneine and its congeners: anti-ageing mechanisms and pharmacophore biosynthesis. Protein & Cell, pages pwad048, 2023.
- 4(1,2,3)
Georges E Janssens, Xin-Xuan Lin, Lluís Millan-Ariño, Alan Kavšek, Ilke Sen, Renée I Seinstra, Nicholas Stroustrup, Ellen AA Nollen, and Christian G Riedel. Transcriptomics-based screening identifies pharmacological inhibition of hsp90 as a means to defer aging. Cell reports, 27(2):467–480, 2019. URL: https://doi.org/10.1016/j.celrep.2019.03.044.
- 5(1,2)
BD Paul and SH Snyder. The unusual amino acid l-ergothioneine is a physiologic cytoprotectant. Cell Death & Differentiation, 17(7):1134–1140, 2010.
